AREST: Arabidopsis Related ESTs
John W. MorrisDepartment of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114. USA
Fax 617-726-6893
email john.morris@frodo.mgh.harvard.edu
With the growing number of EST sequences available through the NCBI's GenBank, it is becoming easier to go prospecting for related genes in other model organisms. The AREST web site was inspired by a presentation at Massachusetts General Hospital by the guest lecturer Dr. Andrea Ballabio on the extensive Drosophila Related EST (DREST) resource. The AREST site like the DREST site, shows blast sequence matches to translated EST sequences. The DREST site shows matches of Drosophila genes against human ESTs, while AREST shows Arabidopsis genes matched against four animal species; human, mouse (Mus musculus), fly (Drosophila melanogaster) and nematode (Caenorhabditus elegans).
Rather than selecting genes with known mutant phenotypes, as in the DREST database, AREST sequences were selected from the all the Arabidopsis sequences in GenBank having both a defined coding sequence (CDS) and a Medline accession. This reduced the approximately 31,000 Arabidopsis sequences in GenBank down to 806. These 806 sequences were matched against the ESTs in GenBank using network tblastn, and the results for the four species above were reformated into HTML for the web display.
The advantage of the AREST web display over the standard blast output is that in the case where several sequences match a particular region of a sequence of interest, all the matches are lined up as a group. One can see clearly the more conserved and variable domains, as shown in the figure below.
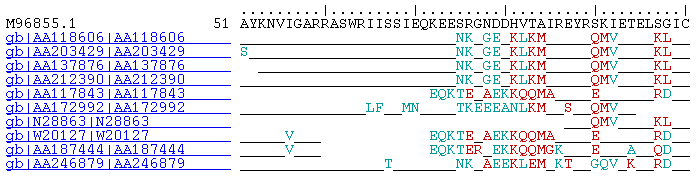
(Exact match = underscore '_', Conservative match = blue-green font, Nonconservative match = red font)
Limitations
1. The AREST displays are meant only to stimulate your interest. They are built from the tblastn reports and so they are unlikely to contain the entire sequence of the match. If you find a set of sequence alignments of interest, download the sequences from GenBank and try the alignment independently before you cut and paste it from the AREST display into your next publication or proposal.
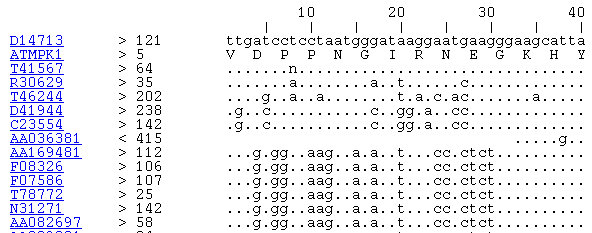
2. Maybe your sequence of interest wasn't included in the set of 806. If this is the case, you can create a somewhat similar display using "power blast" software from NCBI which has a HTML output option. With the help of a web browser, you can view the stacked alignment of matching sequences as shown in the figure below for a blastn alignment. Software is available for Windows, Mac and some UNIX operating systems.
Future Direction
AREST began life as a fun afternoon exercise following a noon seminar by Dr Ballabio, and has grown since then. The hope is that AREST pages will expand to look at human (and mouse?) CDSs matched against Arabidopsis ESTs. Those blast reports may better define some Arabidopsis genes. However, there are hardware and software difficulties to overcome before that happens. So, please visit the site http://weeds.mgh.harvard.edu/arest/ periodically to check on progress.