CAUT home | Background and theory | Working with the CAUT lines Genetics | Generation of the CAUT lines | Seed irradiation and sector analysis
3. Verification of the CAUT lines.
The CAUT lines are phenotypically identical and all are full green and kanamycin resistant. So it is fairly easy to mix them up and ruin your cell-autonomy experiment by using the wrong one. As such errors do happen in any lab a simple and robust method of line verification has been developed. The method is based on Southern blots, T-DNA probes and comparisons to blots run in the Furner lab (see thumbnails below).The parental ch-42 mutant is tagged with a T-DNA containing a promoter less and silent neomycin phosphotransferase and an active hygromycin phosphotransferase conferring hygromycin resistance. The T-DNA also contains a pBR322 copy. Homozygous ch-42 plants are yellow in soil and grow to maturity. Southern blots of DNA from this line prepared with ECORI and probed with pUC18 have a single 9kb fragment corresponding to the tag.
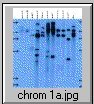
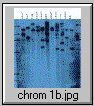
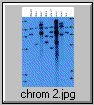
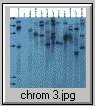
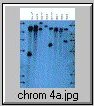
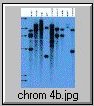
The pCV002GC plasmid used to generate the CAUT lines also has a pBR322 region near the left border (Koncz et al, 1990 The EMBO Journal 9; 1337-1346.). There is one ECORI site in the T-DNA to the right of the pBR322 region. In the transformed plants a second ECORI site is found in the adjacent flanking plant DNA. The second site varies between integration events and therefore between lines. All of the lines are homozygous for a single correction but there can be multiple T-DNA copies at that site. The CAUT lines were made by introducing the correcting T-DNA into the ch-42 mutant. After recurrent back crosses, the single correcting insertions were made homozygous. Southern blots of these lines made with genomic DNA digested with ECORI and probed with pUC18 show a characteristic pattern. All lines have a 9kb band corresponding to the tag at ch-42 on chromosome 4. In addition all lines have one or more bands of varying sizes corresponding to the correcting CH-42 insert at the new location. The fragment pattern is unique to each line and can be used as a distinctive fingerprint to verify the identity of a line.
Below are 7 blots corresponding to the inserts of the 5 chromosomes (chromosomes 1 and 4 are on two blots). The arrows are the positions of lambda HindIII fragments and the sizes (from the top) are: 23.1, 9.4, 6.6, 4.4, 2.3 and 2.0 KB. All lines have the 9kb fragment but there is slight variation in the migration of the fragment.